Introduction
Idiopathic inflammatory myopathies (IIM) belong to the group of systemic autoimmune diseases. Their annual incidence is approximately one in 100,000 people, and they affect more women than men. Regarding clinical symptoms, IIM can be divided into dermatomyositis (DM) and the childhood form juvenile dermatomyositis, polymyositis (PM), inclusion body myositis and necrotizing myopathy [1]. The characteristic symptoms of patients suffering from IIM are muscle weakness, histopathological signs of inflammation in muscle tissues, elevated levels of muscle-associated enzymes in serum, inflammatory mononuclear cells infiltrating muscle tissue and progressive symmetrical proximal muscle weakness [2]. Despite the number of studies focused on identifying the genetic background of IIM, their detailed etiopathogenesis remains unknown [3].
Besides the immunological point of view, a strong association between IIM and major histocompatibility complex (MHC) genes is known. The MHC plays an essential role in T-cell receptor repertoire development, peripheral tolerance to self-antigens and the regulation of many types of immune responses to environmental antigens. The ability to develop specific lymphocyte-mediated immune responses depends on variability in MHC molecules. Loss of control of MHC-mediated function is responsible for the deregulation of self-tolerance and development of autoimmune pathology [4, 5]. HLA association studies have shown the importance of MHC ancestral haplotype 8.1 (HLA-B*08/DRB1*03/DQB1*02/DQA1*05) in autoimmune diseases [3, 6]. Additionally, many studies have shown that more than 40 different autoimmune diseases are susceptible to allelic variations in the HLA region. In many cases, it is not clear whether the susceptibility to the diseases is directly caused by HLA antigens or by other genes located within the MHC region or by a combination of both HLA antigens and other MHC-located genes. Some recent studies have shown that there is an increased occurrence of autoimmune diseases (celiac disease) in patients with IIM in comparison with healthy subjects [7, 8]. These findings additionally support the assumption that HLA polymorphisms may play a dominant role in risk of autoimmunity development in general.
Three heat shock protein 70 (HSP70) genes are located within the MHC, between HLA class I and HLA class II genes. The human MHC-located HSP70s are encoded by three main intronless polymorphic genes in the HLA region: HSPA1L (HSP70-hom), HSPA1A (HSP70-1) and HSPA1B (HSP70-2). Several studies have suggested that HSP70 proteins are involved in the development of not only autoimmune diseases but also of other inflammatory diseases [9–11]. These proteins play a role as central components of the cellular and immune network. They can be divided into 2 groups according to their main functions – intracellular and extracellular proteins. They participate in the cross-presentation of peptides via MHC antigens and are able to stimulate innate and adaptive immunity [12–14]. An additional function of the HSP70 proteins is the blocking of apoptosis in many ways [15].
The objective of this study was to investigate the association between specific polymorphisms within the human MHC and IIM. Genetic polymorphisms within this region are hypothesized to play a crucial role in the initiation of the autoimmune process. By identifying these polymorphisms, we aim to enhance our ability to diagnose and classify the various subtypes of IIM with greater precision.
Material and methods
Study design and participants
We analyzed a cohort of 152 patients suffering from IIM (82 DM and 70 PM; 111 female; 19–85 years of age, median age 60) and 150 healthy controls (69 female; 2–65 years of age, median age 35) from 2012 to 2020. We determined autoantibodies in the IIM group (anti-Ro, anti-Jo-1, anti-PM-Scl, anti-U1RNP, anti-Mi-2, anti-Ku, anti-PL-12, anti-PL-7, anti-EJ, anti-KS, anti-OJ, anti-Zo, anti-SRP, anti-SAE). The size of the cohort of patients was limited by the number of diagnosed patients and it reflects the prevalence of the disease in the Czech Republic. The number of controls analyzed in this study was appropriate for the number of patients. Patients were treated at the Institute of Rheumatology in Prague. The inclusion criteria for patients in this study was the diagnosis determined according to Bohan and Peter criteria [16, 17]. Patients and controls were residents of the central region of the Czech Republic.
Genotyping of HSP70 gene polymorphisms
Genomic DNA was extracted from peripheral blood cells using the Gentra Puregene Blood Kit (cat. no. 158389) (QIAGEN GmbH, Germany). DNA was extracted from freshly collected blood and stored at –70°C prior to DNA analyses. DNA quality and quantity were measured with a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA). The PCR reaction mixture contained 1 µl of forward primer, 1 µl of reverse primer, 6.6 µl of DNase free water, 10 µl of PPP mix, and 0.4 µl of enhancer. The annealing temperatures during amplification differed between genes (64°C for HSPA1A/ HSPA1B genes and 63°C for HSPA1L genes). The PCR product was treated by adding Fast-Exo (cat. no. EN0581, EF065) (Thermo Fisher Scientific, USA) prior to sequencing. For the Sanger sequencing, we used the BigDye Terminator v3.1 Sequencing Kit (cat.no. 4337456) (Thermo Fisher Scientific, USA) and the precipitation was done via the ethanol precipitation method. After ethanol precipitation, each sample was dissolved in Hi-Di Formamide (Thermo Fisher Scientific, USA) and denatured before analysis in the ABI 3130 automated sequencer (Thermo Fisher Scientific, USA).
HLA genotyping
For the HLA genotyping, we used the same cohorts of patients and controls that we used for HSP70 genotyping. We focused only on loci HLA-DRB1 and HLA-DQB1 in our study. The analysis was performed using the PCR-SSO LABType SSO DNA genotyping system (cat. no. RSO2QT, RSO2B1T) (Thermo Fisher Scientific, USA) and Olerup SBT typing kits (cat. no. LG-PD5.2-7(20), AN-PD6.2-3(20)) (CareDx, USA). All HLA typing results were obtained at the medium resolution (four digits) level. Nevertheless, due to the small numbers of samples, the statistical analyses were mainly performed at low resolution DNA typing level.
Statistical analysis
Allelic and genotype frequencies were calculated by direct counting. Differences in allelic and genotype frequencies and their statistical significances were calculated using the commercial software BioEdit 7.0 (https://bioedit.software.informer.com/7.2/) and GraphPad Prism 7.05 (https://www.graphpad.com/). The differences in allelic frequencies were tested with the Fisher exact test and the differences in genotype frequencies were tested with the χ2 test. The p-value < 0.05 was considered significant. When applicable, p-values were corrected for multiple testing (Bonferroni correction).
Results
The HLA-DRB1*03 allele is a pivotal risk factor for idiopathic inflammatory myopathies
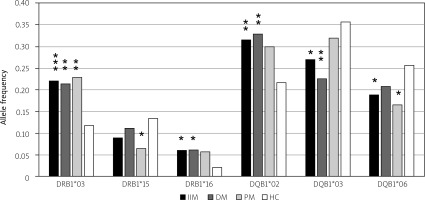
Association of the HLA-DRB1*03 allele with IIM is well documented. We have verified and confirmed this association with DM, PM and IIM (DM and PM together in one group labeled as “IIM”) in Czech patients. Its frequency was elevated in patients suffering from IIM compared to healthy controls, with p < 0.001 (OR = 2.14; 95% CI: 1.37–3.13). We compared the frequency of HLA alleles in patients with DM and PM with healthy controls and found a higher frequency of HLA-DRB1*03 in patients in the DM subgroup, with p < 0.01 (OR = 2.05; 95% CI: 1.22–3.46), and also higher frequency in the PM subgroup, with p < 0.01 (OR = 2.24; 95% CI: 1.33–3.74).
However, the HLA-DRB1*03 allele was not the only significant observation. The frequency of the HLA-DRB1*16 allele was elevated in patients compared to controls, and it was also significantly associated with IIM (p < 0.05; OR = 3.08; 95% CI: 1.28–7.51) and DM (p < 0.05; OR = 3.18; 95% CI: 1.16–8.78). On the other hand, the frequency of the HLA-DRB1*15 allele was lower in patients with PM than in controls. The difference in frequency was statistically significant (p < 0.05; OR = 0.45; 95% CI: 0.21–0.94).
Besides HLA-DRB1 alleles, we analyzed the frequencies of HLA-DQB1 alleles. The frequency of the HLA-DQB1*02 allele was found to be significantly elevated in patients with IIM and in the DM, with p < 0.01 (OR = 1.67; 95% CI: 1.16–2.38) in the IIM group and p < 0.01 (OR = 1.78; 95% CI: 1.15–2.71) in the DM subgroup. We have also found a statistically significant relationship of the HLA-DQB1*03 and HLA-DQB1*06 alleles with IIM. The frequency of the HLA-DQB1*03 allele and the HLA-DQB1*06 allele was lower in patients (IIM, DM) than in the control group. The p-value of the allele frequency difference of the HLA-DQB1*03 allele reached statistical significance in the IIM group (p < 0.05; OR = 0.67; 95% CI: 0.47–0.94) and in the DM subgroup (p < 0.01; OR = 0.53; 95% CI: 0.34–0.80). The frequency of the HLA-DQB1*06 allele was lower in all groups and reached statistical significance in the IIM group (p < 0.05; OR = 0.67; 95% CI: 0.46–0.98) and in the PM group (p < 0.05; OR = 0.57; 95% CI: 0.35–0.95; Fig. 1).
Fig. 1
Statistically significant differences in frequencies of the HLA-DRB1 and the HLA-DQB1 alleles in IIM, DM and PM. We found higher frequencies of the HLA-DRB1*03 allele (p < 0.001), HLA-DRB1*16 allele (p < 0.05), and HLA-DQB1*02 allele (p < 0.01) in patients in comparison with controls. The difference was statistically significant in all tested groups of patients for the HLA-DRB1*03 allele (p < 0.01 in DM, p < 0.01 in PM). The elevated frequency of the HLA-DRB1*16 allele was statistically significant in IIM (p < 0.05) and DM (p < 0.05). Also, the elevated frequency of HLA-DQB1*02 allele was statistically significant (p < 0.01 in IIM, p < 0.01 in DM). On the other hand, we found lower frequencies of the HLA-DRB1*15 allele, HLA-DQB1*03 allele and HLA-DQB1*06 allele in patients compared to controls. The difference was statistically significant for the HLA-DRB1*15 allele in PM (p < 0.05), for the HLA-DQB1*03 allele in IIM (p < 0.05) and DM (p < 0.01), and for the HLA-DQB1*06 allele in IIM (p < 0.05) and PM (p < 0.05).
The polymorphisms of MHC-located HSP70 genes are associated with idiopathic inflammatory myopathies
We analyzed 5 SNPs and one pentanucleotide tandem duplication in HSP70 genes in patients suffering from IIM (DM and PM in one group) and in the cohort of healthy controls. When we compared the allelic frequencies of these polymorphisms between IIM and healthy controls, we found a significantly higher frequency of the C allele (rs1008438) of the HSPA1A gene in patients than in controls (p = 0.01; OR = 1.51; 95% CI: 1.09–2.08) and the G allele (rs1061581) of HSPA1B (p < 0.05; OR = 1.39; 95% CI: 1.00–1.93) in patients with IIM. The frequency of the INS allele of the pentanucleotide tandem duplication AAGTT (rs9281590) in the HSPA1B gene was significantly higher in patients, with p < 0.01 (OR = 1.67; 95% CI: 1.21–2.32). On the other hand, the frequency of the C allele (rs2227956) in the HSPA1L gene was found to be significantly lower in patients than in controls (p = 0.01; OR = 0.52; 95% CI: 0.32–0.84). All statistically significant results are summarized in Table I.
Table I
Summary of statistically significant associations of HSP70 polymorphisms in the patients with IIM and DM
[ii] The comparison of 5 SNPs and one pentanucleotide tandem duplication of HSP70 genes between IIM patients and healthy controls. The frequency of the C allele (rs1008438) of HSPA1A (p = 0.01), the frequency of the G allele (rs1061581) of HSPA1B (p < 0.05) and the frequency of the INS allele (pentanucleotide tandem duplication) (rs9281590) of HSPA1B (p < 0.01) were found to be elevated in patients, with statistical significance. Only the frequency of the C allele of HSPA1L (rs2227956) was found to be lower in patients than in controls (p = 0.01).
[iii] Frequencies of 5 SNPs and one pentanucleotide tandem duplication of HSP70 genes were compared between patients with DM and healthy controls. The frequency of the C allele (rs1008438) of HSPA1A (p < 0.01), the frequency of the C allele (rs1043618) of HSPA1A (p = 0.01), the frequency of the G allele (rs1061581) of HSPA1B (p < 0.05) and the frequency of the INS allele (rs9281590) of HSPA1B (p < 0.01) were found to be elevated in the DM subgroup in comparison with healthy controls. The frequency of the C allele (rs2227956) of HSPA1L was significantly lower in the DM subgroup (p = 0.1).
Subsequently, we excluded all individuals having the known risk allele HLA-DRB1*03 from the IIM group (60 individuals) and the healthy controls group (34 individuals) and compared them to each other. Statistical analysis showed two polymorphisms to be associated with IIM independently of the presence of the HLA-DRB1*03 allele. The frequency of the INS allele of the tandem pentanucleotide duplication AAGTT (rs9281590) located in the HSPA1B gene was higher in patients than in controls (p < 0.05; OR = 1.56; 95% CI: 1.03–2.39). Nevertheless, the frequency of the C allele (rs2227956) located in HSPA1L was found to be lower in patients (p < 0.05; OR = 0.54; 95% CI: 0.31–0.92; Table II).
Table II
HSP70 polymorphisms are associated with the IIM and DM independently of the presence of the HLA-DRB1*03 allele
[ii] After excluding all participants carrying the HLA-DRB1*03 allele, a comparison of five SNPs and one pentanucleotide tandem duplication (AAGTT) of HSP70 genes between patients with IIM and healthy controls was done. The frequency of the INS allele (rs9281590) of HSPA1B was found to be higher in patients (p < 0.05). However, the frequency of the C allele (rs2227956) of HSPA1L was found to be lower in patients (p < 0.05).
In the complete set of DM patients (HLA-DRB1*03 carriers were present, 82 individuals), we found a higher frequency of the C allele (rs1008438) in HSPA1A (p < 0.01; OR = 1.79; 95% CI: 1.22–2.63), a higher frequency of the C allele (rs1043618) in HSPA1A (p = 0.01; OR = 1.69; 95% CI: 1.15–2.48), a higher frequency of the G allele (rs1061581) in HSPA1B (p < 0.05; OR = 1.64; 95% CI: 1.12–2.40), and a higher frequency of the INS allele of the pentanucleotide tandem duplication (rs9281590) in HSPA1B (p < 0.01; OR = 1.82; 95% CI: 1.24–2.70) compared to the complete group of healthy controls (DRB1*03 carriers were present, 150 individuals). Additionally, our results showed a lower frequency of the C allele (rs2227956) in HSPA1L (p = 0.01; OR = 0.45; 95% CI: 0.24–8.83) in DM patients. All mentioned differences reached statistical significance only in the DM subgroup. No statistically significant differences were found in the PM subgroup (72 subjects). The results are shown in Table I.
Subsequently we focused on the HSP70 allele distribution in relation to the presence of the HLA-DRB1*03 allele. We performed the same statistical analysis and frequency comparison, but with exclusion of all individuals carrying the HLA-DRB1*03 allele in the DM subgroup (49 subjects) and in the group of healthy controls (116 subjects). We found a higher frequency of the C allele (rs1008438) in HSPA1A (p < 0.05; OR = 1.74; 95% CI: 1.08–2.86), a higher frequency of the C allele (rs1043618) in HSPA1A (p < 0.05; OR = 1.70; 95% CI: 1.05–2.81) and a higher frequency of the INS allele of the pentanucleotide tandem duplication (rs9281590) in HSPA1B (p < 0.05; OR = 1.86; 95% CI: 1.12–3.03) in the group of patients. These differences reached statistical significance and are summarized in Table II.
Idiopathic inflammatory myopathies are related to the presence of distinct MHC haplotypes
Using Arlequin 3.5 software, we created haplotypes consisting of HLA-DQB1-DRB1-HSP70 genes HSP70 positioned HSPA1L (rs2227956)-HSPA1A (rs1008438)–HSPA1A (rs1043618)-HSPA1B (rs1061581)-HSPA1B (rs539689)-HSPA1B (rs9281590). Haplotypes were created for the group of patients with IIM, DM, PM and healthy controls. We found the strongest association with the elevated frequency of haplotype HLA-DQB1*05-DRB1*16:01-T-A-G-A-G-DEL, with p < 0.001 (OR = 32.18; 95% CI: 1.92–540.60) in IIM, p < 0.001 (OR = 32.64; 95% CI: 1.87–569.60) in the DM subgroup and p < 0.001 (OR = 29.04; 95% CI: 1.62–519.70) in the PM subgroup. Nevertheless, the most frequent haplotype, which had an elevated frequency in the patient group, was HLA-DQB1*02-DRB1*03:01-T-C-C-G-C-INS, with p < 0.05 (OR = 1.90; 95% CI: 1.15–3.13) in IIM and p < 0.05 (OR = 1.82; 95% CI: 1.02–3.25) in the DM subgroup. The summary of all haplotypes with p-values are shown in Table III.
Table III
Comparison of MHC haplotypes between patients with IIM, DM, PM and healthy controls
[i] DM – dermatomyositis, IIM – idiopathic inflammatory myopathy, ns – non-significant, PM – polymyositis.
[ii] The frequencies of haplotypes consisting of HLA-DQB1-DRB1-HSP70 genes positioned at HSPA1L (rs2227956)-HSPA1A (rs1008438)-HSPA1A (rs1043618)-HSPA1B (rs1061581)-HSPA1B (rs539689)-HSPA1B (rs9281590) were compared between all patient groups (IIM, DM, PM) and controls. The strongest association with disease was found for the HLA-DQB1*05-DRB1*16:01-T-A-G-A-G-DEL haplotype (elevated frequencies in IIM, DM, PM) (p < 0.0001). The most frequent haplotype among all patient groups was the HLA-DQB1*02-DRB1*03:01-T-C-C-G-C-INS haplotype, with p < 0.05.
Haplotype HLA-DQB1*02-DRB1*03:01 has the strongest association with idiopathic inflammatory myopathies
Idiopathic inflammatory myopathies are related to the presence of distinct HLA haplotypes. We counted the frequencies of identified haplotypes in the IIM group consisting only of HLA-DRB1 and DQB1 alleles (polymorphisms of HSP70 genes were not included). The most frequent and significantly associated haplotype with IIM was HLA-DQB1*02–DRB1*03:01 (p < 0.001; OR = 2.36; 95% CI: 1.49–3.75).
Autoantibodies
Autoantibodies related to IIM are normally found in more than 50% of patients. They are known as myositis-specific autoantibodies (MSAs) and myositis-associated autoantibodies (MAAs), depending on related conditions [18]. Myositis-specific autoantibodies are associated with specific phenotypes of skin, muscle, lung disease and malignancy. On the other hand, MAAs are also found in other systemic autoimmune rheumatic diseases. Detection of all these autoantibodies leads to better understanding, diagnosis, classification and treatment of IIM [19].
We also tested the presence of autoantibodies in the group of IIM patients. For the measurement of all autoantibodies, commercially available ELISA tests were used. In our group of 46 analyzed patients, at least one type of following autoantibodies was found: anti-Ro, anti- Jo-1, anti-PM-Scl, anti-U1RNP, anti-Mi-2, anti-Ku, anti- PL-12, anti-PL-7, anti-EJ, anti-KS, anti-OJ, anti-Zo, anti-SRP, anti-SAE. The most frequent autoantibody was anti- Jo-1, with the frequency of 11.84%, followed by anti-PM-SCL, with the frequency of 11.18%, and anti-Ro, with the frequency of 8.55%. Due to the small number of patients positive for presence of autoantibodies, we did not perform any further analysis. More detailed demographic information about patients and information about autoantibodies are given in Table IV.
Table IV
Demographic data of patients with IIM
Discussion
An increasing frequency of patients suffering from autoimmune diseases such as diabetes mellitus, rheumatic arthritis, lupus erythematosus, celiac disease, etc. has been seen in recent years (see American Autoimmune Related Diseases Association reports). Idiopathic inflammatory myopathy belongs to the group of autoimmune diseases where the association with the ancestral haplotype HLA-DRB1*03-DQB1*02 is known [20, 21]. We confirmed that both of these alleles were significantly associated with the disease in our group of subjects suffering from IIM. Surprisingly, we also found a new, statistically significant association of the HLA-DRB1*16 allele in patients with IIM and DM. On the other hand, the frequency of the HLA-DRB1*15 allele was lower in the PM subgroup compared to healthy controls, suggesting that it may have a protective effect.
Presence of the HLA-DRB1*16 allele is not typical for IIM as it appears in low frequencies in the European population and in the Czech Republic as well. The frequency of the HLA-DRB1*16 allele in the tested groups was 5.9% in patients with IIM, 6.1% in patients with DM, 5.7% in patients with PM and 2% in healthy controls. Recent information published on the Allele Frequency Net Database website shows the frequency of 3.8% of the HLA-DRB1*16 allele in the Czech population [22]. The frequency of the DRB1*16 allele was significantly elevated in patients suffering from heterogeneous autoimmune disease myasthenia gravis in the Italian population, as reported by Testi et al. [23–25].
Analysis of the HLA-DRB1*15 allele frequency shows an interesting result pointing to the fact that this allele can have a protective role for PM development. This allele is known to be in linkage disequilibrium with HLA-DQB1*06 [26]. The DQB1*06 allele was found to be protective for PM in our study. The frequency of the HLA-DRB1*15 allele was lower in PM, only 6.4 %, compared to 13% in the healthy controls. Our findings are supported by the authors Bettencourt et al. [27], who reported evidence for the protective role of HLA-DRB1*13 and HLA-DRB1*15 against autoimmune systemic sclerosis development. Moreover, the HLA genotyping for PM/DM patients was performed by Flam et al. [28], and the protective role of the HLA-DRB1*15 allele against autoimmune disease has been shown as well [29].
In our study, we found different associations of HLA alleles with PM and with DM. HLA-DRB1*16 has a predisposing role for DM, and HLA-DRB1*15 plays a protective role in PM. According to these differences, we can confirm that DM and PM have different genetic backgrounds.
Idiopathic inflammatory myopathy is associated with the ancestral haplotype HLA-DRB1*03-DQB1*02, and we observed a significant association for both of these alleles. In addition, we found the HLA-DQB1*03 and the HLA-DQB1*06 allele to have a significant protective effect on IIM development. In diabetes mellitus (including latent autoimmune diabetes), the majority of studies have identified the protective function of the DQB1*06 allele against the disease [29–31]. The HLA-DQB1*03 allele is in linkage disequilibrium with HLA-DRB1*04 and HLA-DRB1*11 [26]. Our data suggest a potential protective effect of both of these alleles on IIM, but our results were not statistically significant.
Several studies have supported the hypothesis that polymorphisms in the heat shock protein 70 gene may be linked with multiple autoimmune disorders. Such an association is often attributed to the development of distinct MHC haplotypes, which arise due to an imbalance between alleles in this genomic region. Therefore, we hypothesized that these polymorphisms might also be involved in the etiopathogenesis of autoimmunity. Our data showed a significant relationship between IIM and the C allele (rs1008438) of HSPA1A, the G allele (rs1061581) of HSPA1B, the C allele (rs2227956) of HSPA1L and the insertion of AAGTT (rs9281590) of the HSPA1B gene. All significantly associated alleles have a predisposing effect on IIM except the C allele (rs2227956) of the HSPA1L gene. This allele has a protective effect. Studies on polymorphisms of HSP70 have shown that these polymorphisms of HSP70 genes are associated with autoimmune diseases, such as inflammatory bowel disease, sarcoidosis, spondyloarthropathies and other diseases [32–34]. However, no articles describing the insertion/deletion rs9281590 and rs1061581 of the HSPA1B gene and rs1061581 of HSPA1B in relation to autoimmune diseases have been published so far.
The HLA-DRB1*03 allele is known for its strong association with autoimmune diseases in general. Therefore, we excluded all healthy controls and patients (the DM group together with PM) carrying the HLA-DRB1*03 allele and we performed the same comparison of 5 SNPs and one pentanucleotide tandem duplication of HSP70 genes. Our results showed that the absence of the DRB1*03 allele does not affect the importance and relationship of some HSP70 polymorphisms with the development of a disease. Allele frequencies of the C allele (rs2227957) of the HSPA1L gene and the pentanucleotide tandem duplication AAGTT (rs9281590) of the HSPA1B gene remained significantly different between the controls and the patients. The C allele (rs2227957) of HSPA1L retained its protective effect on IIM.
Polymyositis is an inflammatory myopathy mediated by cytotoxic T cells, while DM is an autoantibody-mediated angiopathy resulting in typical dermatitis [35]. The different clinical symptoms of PM lead to an assumption of a different genetic background in comparison to DM. This assumption was repeatedly confirmed by our results. We compared 5 SNP polymorphisms and a pentanucleotide tandem duplication of HSP70 genes of the patients with DM and the healthy controls. The differences in frequencies of the C allele (rs2227956) of HSPA1L, the C allele (rs 1008438) of HSPA1A, the C allele (rs1043618) of HSPA1A, the G allele (rs 1061581) of HSPA1B and the pentanucleotide tandem duplication AAGTT (rs9281590) of HSPA1B were significant. The C allele (rs2227956) of HSPA1L has retained its protective function compared to other SNPs of HSP70 genes. We did not find any statistically significant results for comparison of PM with the healthy controls.
The HLA-DRB1*03 allele has a strong effect on autoimmunity, as discussed above. Therefore, we eliminated this effect by excluding the DRB1*03 positive individuals (only DM patients and controls) from the analysis. We observed statistically significant differences in allele distribution of the C allele (rs1008438) of HSPA1A, the C allele (rs1043618) of HSPA1A and the insertion of the pentanucleotide tandem duplication AAGTT (rs9281590) of HSPA1B. We performed similar comparisons in patients with PM and we did not find any statistically significant results. Based on our findings, significant differences were found in the genetic background between DM and PM.
HLA genes are close to each other, are in linkage disequilibrium and are inherited in certain blocks. HSP70 genes are located within the MHC, particularly between HLA class I and class II; therefore, HSP70 genes can be inherited in strong linkage together with the HLA alleles. Using Arlequin software and subsequent statistical analyses, we found the most statistically significant haplotype. It was HLA-DQB1*05-DRB1*16:01-T-A-G-A-G-DEL, with p < 0.001 for IIM, DM and PM groups. Nevertheless, the HLA-DRB1*16:01 allele appeared in low frequency alone, together with DQB1*05 and 5 SNPs and one pentanucleotide tandem duplication AAGTT of HSP70 appeared only in 15 cases in IIM and in none of the healthy controls. The most frequent haplotype in IIM and DM was HLA-DQB1*02-DRB1*03:01-T-C-C-G-C-INS, with p < 0.05. It seems that the most frequent and statistically significant haplotype, HLA-DQB1*02-DRB1*03:01-T-C-C-G-C-INS, could be specific for subjects with IIM or DM respectively (this haplotype appeared in 15.8% in IIM; 15.2% in DM). The most frequent haplotype for PM is HLA-DQB1*02-DRB1*03:01-T-C-C-G-C-INS. The second most frequent and statistically significant haplotype specific for PM is HLA-DQB1*06-DRB1*15:01-C-A-G-A-C-DEL.
It is commonly understood that HLA alleles associated with diseases are linked to the presence of disease-specific or disease-associated autoantibodies. In the study performed by Chinoy et al. [36], a strong association in IIM was observed between HLA-DRB1*03 and anti-Jo-1 status. Additionally, in African American patients, there was a significant correlation between the frequency of the DQA1*01:02 allele and the anti-signal recognition particle (anti-SRP), while the DRB1*03:02 allele was associated with anti-Mi-2 autoantibodies [37, 38]. Furthermore, DRB1*04:05 was found to be elevated in Japanese patients with anti-ARS autoantibodies compared to the controls [38]. Many studies have mentioned that immune-mediated myositis (IIM) is associated with various kinds of autoantibodies. However, we should mention that our analysis was limited by insufficient data on these antibodies, which precludes us from conducting statistical analyses due to small sample sizes.
Conclusions
This study investigated the link between specific MHC polymorphisms and IIM. Genetic variants in this region are critical to autoimmune processes, influencing IIM subtype diagnosis and classification. The findings highlight HSP70 polymorphisms and HLA-associated alleles as key factors in IIM pathogenesis, offering insights for improved diagnostics and therapies. However, further research across diverse populations is needed to confirm these results.